Screening and Potential of Gram Negative Bacterial Isolates for their Extracellular Enzymatic Activities Isolated from the Hospital Aquatic Environment
Citation: Pharmaceutics, Industrial pharmacy, Medicinal chemistry,
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@jbclinpharm.org
Abstract
Bacteria can resist an antimicrobial agent by producing extracellular enzymes that eradicate or inactivate the antibiotics. Majority of drug/metal resistant Gram negative bacterial isolates showed amylase, β-Lactamase, protease, lipase, gelatinase and urease enzymatic activity. In isolates from site-1, maximum 96%, 74% and 72% of the total isolates showed amylase, lipase and catalase activity, whereas only 46%, 34%, 24% and 20% exhibited β-Lactamase, urease, protease and gelatinase activity respectively. In site-II, 88%, 84%, 74%, 68%, 58% and 56% of the isolates demonstrated lipase, protease, amylase, catalase, gelatinase and urease activity respectively, while only 48% of the isolates showed β-Lactamase activity. In case of site- III, maximum 86%, 74%, 68%, 62%, 56% and 52% of the total isolates exhibited lipase, amylase, β-Lactamase, catalase, urease and protease activity respectively. Whereas, only 46% of the isolates demonstrated gelatinase activity. In case of site-IV, maximum 96%, 80%, 66%, 62% of the isolates showed catalase, protease, β-Lactamase and gelatinase activity respectively. Whereas only 48% and 46% of the isolates demonstrated amylase, lipase and urease activity. In our observation most of the isolates from site I, II, III showing lipase activity its mean the hospital wastewater have been suitable source (carbon, nitrogen, salt and mineral) for growth and production of lipase activity. This result revealed that the presence of multi drug/metal resistance Gram negative bacteria from the hospital wastewater were showing various amylase, protease, lipase, catalase, betalactamase, gelatinase and urease, resistance mechanisms and this drug resistant strains may cause infections to the healthy living things. To reduce the spread of drug/metal resistant isolates from hospital to environment, and use good safety sterilization methods before release of waste materials to the environment or sewage.
Keywords
Drug resistance; extracellular enzymes; hospital wastewater; gram negative bacteria
Introduction
Bacteria can resist an antimicrobial agent by producing extracellular enzymes that eliminate or inactivate the antibiotics. As an example, penicillins, monobactams, carbapenems, and cephalosporin’s are identified chemically as beta-lactam antibiotics, and many bacteria happen to resistant to these drugs by producing various beta-lactamases that are capable to inactivate some forms of these antibiotics. Beta-lactamases break the beta-lactam ring of the antibiotics, thus demolishing the antimicrobials. Bacteria may develop resistant to aminoglycosides (Streptomycin, netilmicin, neomycin, gentamicin, tobramycin, amikacin, etc.) by enzymatically adding new chemical groups to these antibiotics, thus inactivating the antibiotics. In Gram-negative bacteria, the antibiotics inactivating enzymes are frequently situated in the periplasmic space placed between the peptidoglycan layers and the external membrane and interact with the antimicrobials. Gram-positive bacteria not have an external membrane and the antimicrobial inactivating enzymes are typically secreted from the bacteria and interact with the antibiotics. Amylases are starch degrading enzymes. They are extensively spread in microorganism, plants and animals kingdoms. They degrade starch and associated polymers to yield products quality of individual amylolytic enzymes. [1] Tallur[2] characterized bacterial isolates, Bacillus toyonensis PNTB1, Lysinibacillus sphaericus PTB, Vibrio vulnificus PMD, Shewanella MPTDBS, and Pseudomonas chlororaphis PNTB for their tolerance to metal and antibiotics. Vibrio vulnificus PMD demonstrate maximum tolerance at higher concentration of metal than other bacteria. These bacterial isolates were test for the production of extracellular enzymes such as lipase, pectinase, tannase, cellulase, chitinase, and l-glutaminase. Vibrio vulnificus exhibited higher production of l-glutaminase enzyme. Bacillus toyonensis PNTB1 demonstrate lipase, CM-cellulase and chitinase activities. Baral[3] reported that the extended spectrum b-lactamase (ESBL) production was detected in 55.2% of a subset of MDR E. coli isolates. Rahman[4] screened extracellular enzymes (amylase, cellulase and xylanase) among the 52 bacterial isolates from the soil and water samples. Four isolates were found to be positive for amylase, 10 were found positive for xylanase, 7 were detected positive for pullulanase and 2 for cellulase activities. Ong[5] studied enzymatic test in E. coli and reported that the all isolates were proteolytic phosphatase and lipolytic enzyme producers. Amylolytic activity did not exist in all isolates. Protease, leucine arylamidase, alkaline and acid phosphatases, esterase lipase and phospholipase esterase were find out in all the isolates. In this study, we isolate and identify the Gram negative bacteria from the different hospital wastewater. Furthermore, the antibiotic sensitivity and extracellular enzymes activity were characterized. The results will clarify the phenotypic characteristics and, especially, the virulent traits of Gram negative bacteria to provide the theoretical basis in disease control.
Material and Methods
Sampling and sampling sites
Water samples were collected from three sites of hospital wastewater along with King George’s Medical University Site (I), Sanjay Gandhi Post Graduate Institute of Medical Sciences (treated) Site (II), Sanjay Gandhi Post Graduate Institute of Medical Sciences (untreated) Site (III) and Dr. Ram Manohar Lohia Hospital Site (IV) at Lucknow city as shown in Figure 1. Samples were collected in sterile 250-ml polypropylene bottles, according to internationally recommended methodology.[6] Samples were kept at 4°C until their arrival to laboratory.
Isolation and identification of metal tolerant gram negative bacteria
Isolation of metal resistant Gram negative bacteria from wastewater samples were done on metal amended Mac conky agar plates at varying concentration (25-1200 µg/ml). Serial dilutions of the wastewater samples were plated by spreading 0.1 ml on medium for the count of total metal resistant Gram negative bacteria. Plates incubated at 37°C for 24 hours and Gram negative bacterial counts including pink and colorless colonies were expressed as CFU/ml on Mac conky agar medium. These isolates were finally identified on the basis of morphological, cultural and biochemical characteristics including IMVic tests (indole, methyl red, voges proskeur and citrate utilization tests, carbohydrate test, hydrogen sulphid test etc.[7]
Determination of antibiotic resistance
The antibiotic resistance was determined by a standard disc diffusion technique using Mueller-Hinton agar (Difco) according to the recommendations of National Committee for Clinical Laboratory Standards (NCCLS 2008) including Escherichia coli ATCC 25922 as a control strain. The antimicrobial drugs tested and their sensidisk concentrations were: Amoxicillin (AMX) 25 µg, Nalidixic acid (NA) 30 µg, Neomycin (N) 30 µg, Kanamycin (K) 30 µg, Ampicillin (AMP) 10 µg, Gentamycin (GEN) 30 µg, Nitrofurazole (NR) 100 µg, Chloramphenicol (CHL) 30 µg, Polymixine B (PB) 300 µg, Methicillin (M) 5 µg, Penicillin (P) 10 µg, Ciprofloxacin (CIP) 5 µg, Erythromycin (ERY) 15 µg, Oflaxicin (OF) 5 µg and Tetracycline (TET) 10 µg. Within 15 min of the application of the discs, the plates were inverted and incubated at 37°C. After 24 h of incubation, the plates were examined, and the diameters of the zones of complete inhibition to the nearest whole millimetre were measured. The zone diameter for individual antimicrobial agents was then translated into sensitive and resistant categories.[8]
Beta lactamase production
For detection of beta lactamase producing bacteria a loop full of grown culture will be transferred into small tube containing 1 ml of penicillin G solution and incubated at 37°C for 30 min. 0.5 ml of iodine solution will be added and mixed for 2-3 min. Change in colour to colourless, indicates as positive result.[9]
Amylase test
Starch degrading activity of the cultures was screened by hydrolysis of starch on a medium (g/l) containing 5 g peptone, 3 g beef extract, 2 g starch (soluble), and 20 g agar. Overnight Gram negative bacterial growth on Petri dishes was flooded with an iodine solution.[9] Development of a pale yellow zone around a colony in an otherwise blue medium has indicated starch degrading activity. The isolates were considered positive for amylase production.
Protease test
Protease producing enzymatic activity of the cultures was screened; single streak inoculation of the test microbe was done on agar surface. Incubate the plates at 37°C for 24-48 hours in an inverted position. Presence of clear area around the line of growth was observed, indicates the positive result.
Lipase test
Gram negative bacterial isolates were inoculated on tributyrin agar medium (g/l) containing 5 g peptone, 3 g beef extract, 10 tributyrin and 15 g agar, pH 6.5. After incubation at 28°C for 7 days the development of a clear halo zone around the colony indicated lipase activity.[10]
Catalase test
Catalase producing enzymatic activity of the cultures was screened; trypticase soy agar medium slants were prepared. Inoculation of test microbe was done and slants were kept at 37°C for 24-48 hours. Then, pour 3-4 drops of H2O2 gas bubbles were released it shows the positive result.
Gelatinase test
Test to detect gelatin hydrolysis is the Nutrient Gelatin plate method. In this method, heavy inoculums of an 18-24 hour old test bacteria is stab-inoculated onto culture plates pre-filled with Nutrient Gelatin (23 g/L Nutrient Agar; 8 g/L 275 bloom gelatin). Inoculated Nutrient Gelatin plates are incubated at 35°C for 24 hours. Gelatin hydrolysis is indicated by clear zones around Gelatinase-positive colonies. In some cases, plates are flooded with mercuric chloride solution to precipitate unhydrolyzed gelatin making the clear zones easier to see. Results are often observed within 5-10 minutes after flooding with mercuric chloride solution.[11]
Urease test
Agar plate containing peptone 1.0 g (g/L), sodium chloride 5.0 g (g/L), potassium monohydrogen (or dihydrogen) phosphate 2.0 g (g/L), agar 20 g (g/L). Heat to boil and to dissolve the medium completely, adjust the pH to 6.8. Sterilize by autoclaving at 15 lbs pressure (121°C) for 15 minutes and cool it to 50°C, then add glucose 1.0 g and phenol red (0.2% solution) 6.0 ml and add molten base and steam for I hour, cool to 50°C add 20% aqueous solution 100.0 ml mix well and distribute into sterile containers. Examine the slants/broth as their color for the presence of urease (red color) and non-urease (yellow color).[11]
Results
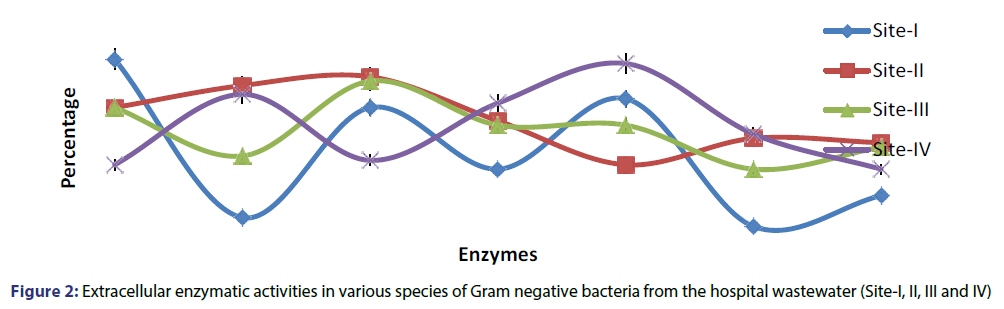
The heavy metal and antibiotics resistance among the Gram negative bacterial population varied considerably in different metal/ antibiotic and water sampling sites. Gram negative bacteria showed lower metal resistant viable count range 4.01 × 104-1.3 × 103 at 50-100 µg/ml in site-IV as compared to 11.03 × 105-1.03 × 104, 12.02 × 105-1.4 × 103 and 12.33 × 105-2.7 × 103 in site-I, II and III against all metal tested, respectively. Of the 200 Gram negative bacterial isolates tested, marked antibiotic resistances (over 90%) were observed for amoxicillin, methicillin, ampicillin, nalidixic acid and penicillin depending upon the sampling sites. All Gram negative bacterial isolates also showed multiple resistance patterns (2-13 antibiotics) in different combination of antibiotics. Majority of drug/metal resistant Gram negative bacterial isolates showed amylase, ß-Lactamase, protease, lipase, gelatinase and urease enzymatic activity. In isolates from site-1, maximum 96%, 74% and 72% of the total isolates showed amylase, lipase and catalase activity, whereas only 46%, 34%, 24% and 20% exhibited ß-Lactamase, urease, protease and gelatinase activity respectively. In site-II, 88%, 84%, 74%, 68%, 58% and 56% of the isolates demonstrated lipase, protease, amylase, catalase, gelatinase and urease activity respectively, while only 48% of the isolates showed ß-Lactamase activity. In case of site- III, maximum 86%, 74%, 68%, 62%, 56% and 52% of the total isolates exhibited lipase, amylase, ß-Lactamase, catalase, urease and protease activity respectively. Whereas, only 46% of the isolates demonstrated gelatinase activity. In case of site-IV, maximum 96%, 80%, 66%, 62% of the isolates showed catalase, protease, ß-Lactamase and gelatinase activity respectively. Whereas only 48% and 46% of the isolates demonstrated amylase, lipase and urease activity. Maximum 74.5%, 73% and 60% of the isolates showed catalase, amylase and lipase and protease activity, whereas, only 45% isolates showed urease activity from all the sites tested respectively [Figure 2].
Species based screening of extra cellular enzymatic activities were also determined from entire sites collectively. Majority of Gram negative bacteria including Proteus vulgaris, E. coli, Enterobacter aerogenes, Pseudomonas aeruginosa, Klebsiella pneumoniae, Salmonella typhi, Citrobacter amalonaticus and Serratia marcescens isolates produced amylase, ß-Lactamase, protease, lipase, gelatinase and urease enzymatic activity. In Proteus vulgaris Maximum, 98%, 84% and 80% of the total isolates showed urease, gelatinase and amylase and lipase activity respectively. 71.1% and 60% of E. coli isolates demonstrated catalase and amylase and ß-Lactamase activity, while 77.4% and 67.7%, of Enterobacter aerogenes isolates exhibited catalase, amylase and lipase activity, respectively. Maximum 82.1% and 78.5 of the Pseudomonas aeruginosa showed lipase and catalase activity respectively. While 92.3%, 88.4% and 76.9% of the Klebsiella pneumoniae showed urease, gelatinase and lipase activity respectively. In case of Salmonella typhi maximum, 63.6% of isolates exhibited amylase activity. While in case of Citrobacter amalonaticus, 81.8% isolates showed ß-Lactamase and lipase activity. 100% of the Serratia marcescens showed lipase and gelatinase activity [Table 1].
| S No | ß-Lactamase | Lipases | Proteases | Amylases | Catalase | Gelatinase | Urease |
|---|---|---|---|---|---|---|---|
| Proteus vulgaris | 56% | 80% | 68% | 80% | 74% | 84% | 98% |
| E. coli | 60% | 57.1% | 54.2% | 60% | 77.1% | 14.2% | 22.8% |
| Enterobactor aerogenes | 51.6% | 67.7% | 45.1% | 77.4% | 77.4% | 6.4% | 9.6% |
| Pseudomonas aeruginosa | 57.1% | 82.1% | 57.1% | 75% | 78.5% | 10.7% | 0% |
| Klebsiella pneumoniae | 65.3% | 76.9% | 57.6% | 69.2% | 61.5% | 88.4% | 92.3% |
| Salmonella typhii | 18.1% | 27.2% | 27.2.5% | 63.6% | 18.1% | 0% | 9.0% |
| Citrobacter amalonaticus | 81.8% | 81.8% | 72.7% | 63.6% | 81.8% | 45.4% | 27.2% |
| Serratia marcescens | 50% | 100% | 87.5% | 75% | 87.5% | 100% | 50% |
Table 1: Extracellular enzymatic activities in various species of Gram negative bacteria from the hospital wastewater (Proteus vulgaris, E. coli, Enterobacter aerogenes and Pseudomonas aeruginosa, Klebsiella pneumoniae, Salmonella typhi, Citrobacter amalonaticus and Serratia marcescens).
Discussion
Environmental pollution is one of the most serious problem of the modern time specially water pollution in India due to the dangerously high levels of antibiotics/metal and other pollutants. Studies have shown that the release of wastewater from hospitals was associated with an increase in the prevalence of antibiotic resistance. Production of extracellular photolytic enzymes is a property shared by non-infectious and infectious microorganisms. These enzymes are crucial factors in their life cycles and may be mortal to the host when created by infectious bacteria.[12] The role of proteases in pathogenesis is not defined, but it appeard that they are concerned in colonization and offensive during host-infection interaction, apart from provide nutrients for the microorganism. Nailah[13] reported that all the bacterial isolates produces amylase and protease, while only few strain of Gram negative bacteria could produce lipase enzymes. Bacteria produces over 1300 diverse enzymes called ß-lactamases which exist as four classes: A, B, C and D. Class D ß-lactamase enzymes, also known as oxacillinases or OXA enzymes, are able of deactivating a broad range of ß-lactam antibiotics, including “last resort†drugs such as the carbapenems. OXA carbapenemases are mainly find out in Gram-negative bacteria and are potentially contributory to the resistance of carbapenems in clinical isolates. Their host species include Acinetobacter baumannii and A pittii, which are becoming vital human infections due generally to the incident of ß-lactam resistance. Carbapenem resistance has enlarged due to the multiply of such OXA type ß-lactamases.[14,15] Carbapenemase-producing Enterobacteriaceae (CPE) are Gramnegative bacteria that are nearly resistant to the last line Carbapenem class of antibiotics and have appeared in the UK over the past decade. Bacterium may also attain resistance to antimicrobial agents during some mechanisms including production of ß-lactamases, up regulation of efflux pumps, and alteration or down regulation of outer membrane porins.[16] In our observation, maximum 74.5%, 73% and 60% of the isolates showed catalase, amylase, lipase and protease activity whereas only 45% isolates showed urease activity from all the sites tested respectively. Our study supported by some other researcher.[17-19] In our observation most of the isolates from site-I, II, III showing lipase activity its mean the hospital wastewater have been suitable source (carbon, nitrogen, salt and mineral) for growth and production of lipase activity. Similar study reported by others.[20,21] In our observation maximum number of Gram negative bacterial isolates produces amylase, betalactamase, protease and lipase while just few bacteria could produce gelatinase and urease enzymatic activity. Similar study also reported by many other workers.[22,23] Nicoloff [24] confirmed IR in the existence of ß-lactam antibiotics (mecillinam and ampicillin). They examined that the bacteria rapid antibiotic-modifying or -degrading enzymes Ere(A), Tet(X2) or CatA1 caused IR in the incidence of macrolides (clarithromycin and erythromycin), tetracycline’s (tigecycline and tetracycline) and chloramphenicol respectively. Rahman (3) screened extracellular enzymes (xylanase, amylase and cellulase) among the 52 isolates from various water and soil samples. Four isolates were found to be positive for amylase, 10 were positive for xylanase, 7 were positive for pullulanase and 2 for cellulase activities. Representative isolates was preliminarily known genera of Anoxybacillus, Geobacillus and Bacillus sp. The results demonstrate that hot springs in Malaysia are rich in microbial life from the Bacillaceae family, and are dynamic as a source of biocatalysts. Albertson[25] reported that the aquatic bacteria produce many extracellular enzymes. Isolates 59 and a Vibrio sp. Isolates S14 increase extracellular enzyme production under them examined DNase, lipase, chitinase, amylase, and several proteases. The Sl4 isolates produces a DNase, chitinase, lipase and proreases between isolates. Biswas[26] screened Halococc for the production of extracellular enzymes such as glutaminase, amylase, aspa raginase, gelatinase, inulinase, xylanase, cellulasecaseinase, pectinase, urease and lipase. Among these hydrolyt ic enzymes, glutamine and asparagine hydrolytic activities were main, although lipid and casein degrading activities were not lesser. However, amylase and gelatinase production were uncommon. None of the halophiles was competent to degrade cellulose, inulin, pectin and xylan and only one isolate was capable of hydrolyzing urea. Ong (5) studied enzymatic test in E. coli and reported that the all isolates were proteolytic phosphatase and lipolytic enzyme producers. Amylolytic activity did not exist in all isolates. Protease, leucine arylamidase, alkaline and acid phosphatases, esterase lipase and phospholipase esterase were find out in all the isolates. Khalil[27] screened clinical isolates for their extracellular activities. OF 95.2% isolates showed haemolytic activity followed by 83.7%, 81% and 73.3% protease, lipases and gelatinase, respectively. Our findings were also in agreement with other reports[28,29] that the role of enzymes in antibiotic resistance is valuable. Vila[30] reported that the antibiotic resistance is mostly relevant in infectious Aeromonas species in which, besides the classical resistance to ß-lactam antibiotics, multi resistance has been usually identified. Baral[4] reported that the extended spectrum b-lactamase (ESBL) production was detected in 55.2% of a subset of MDR E. coli isolates. John et al. studied Aeromonas sp for their extra cellular enzymatic activities. All the isolates from both fish and water samples produced gelatinase and nuclease but the capability to produce lipase, caseinase and haemolysins was found to be different among isolates from various samples. Among the 15 antibiotics to which the isolates were tested, all the isolates were found to be sensitive to ciprofloxacin, gentamicin and chloramphenicol and resistant to amoxycillin. Present study revealed that the presence of multi drug/ metal resistance Gram negative bacteria from the hospital wastewater were showing various amylase, protease, lipase, catalase, betalactamase, gelatinase and urease, resistance mechanisms and this drug resistant strains may cause infections to the healthy living things.[31]
Conclusion
In our work we analyzed the enzymatic activity among metal tolerance and antibiotic resistance bacteria from hospital effluents. Bacterial pigmentation and enzymatic activities such as, protease, amylase, lipase, betalactamase, catalase, gelatinase and urease were the characteristics chosen to study these effects. We find it relevant to evaluate the possible interactions between antibiotic and metal tolerant bacteria in regard to the physiological and metabolic alterations derived from this contact. We suggest to minimize the spread of drug resistant isolates from hospital to environment is crucial, so good safety sterilization methods to be adopted before release of waste materials to the environment or sewage.
Competing Interest
The authors declare that there are no competing interests.
Author Contribution
Each author should have participated sufficiently in the work to take public responsibility for appropriate portions of the content. Collection of sample, experimental work and supervision of the research group. Manzar Alam carried out the all experimental studies and drafted the manuscript, Mohd Imran participated in the design and oversee the study. All authors read and approved the final manuscript.
Acknowledgements
We are thankful to Prof. SW Akhtar, Vice Chancellor, Integral University, for providing the necessary facility to conduct this research. The authors also thank the HOD, Department of Biosciences and Bioengineering, Integral University, Lucknow for guidance and their cooperation with regard the research work.
REFERENCES
- Burhan A, Nisa U, Gokhan C. Enzymatic properties of a novel thermophilic, alkaline and chelator resistant amylase from an alkalophilic Bacillus sp. Isolate ANT-6. Process Biochem 2003;38:1397-1403.
- Preeti NT, Dayanand BS, Sikandar IM. Characterization of antibiotic resistant and enzyme producing bacterial strains isolated from the Arabian Sea. Biotech 2016;6:28.
- Rehman A, Shakoori AR. Heavy metal resistance Chlorella sp, isolated from tannery effluents, and their role in remediation of hexavalent chromium in industrial waste water. Bulletin of Environmental Contamination and Toxicology 2001;66:542-7.
- Baral P, Neupane S, Marasini BP. Prevalence of multidrug resistance in bacterial uropathogens from Kathmandu, Nepal. BMC Res 2012;5:38.
- Ong KY, Chin HS, Teo KC. Biological screening of microbes isolated from soil of ex-tin mining land in Kampar Area. African J Microbiol Res 2011;5:4757-63.
- Lösch LS, Alonso JM, Merino LA. Occurrence of antimicrobial-resistant Enterobacteriaceae in water from different sources in a subtropical region of Argentina. Revista Ambi Agua, Taubate 2008;2:28-36.
- Cappuccino JG, Sherman N. Microbiology lab manual. USA, Benjamin-Cummings Publishing Company 1995;477.
- Florea AB. Antimicrobial susceptibility of E. coli isolated from aries river (Romania). Analele Universitatii din Oradea - Fascicula Biologie 2011;18:34-8.
- Barry AL, Bernsohn AL, Adams AP. Improved 18-hour methyl red test. Applied Microbiol 1970;20:866-70.
- Cappuccino JG, Sherman N. Microbiology - A laboratory manual 1996;159-201.
- Aneja KR. Experiments in Microbiology Plant Pathology and Biotechnology. New Age International Pvt. Ltd., Publishers, New Delhi, India 2003. 4th edn.
- Miyoshi S, Shinoda S. Microbial metalloproteases and pathogenesis. Microbes Infect 2000;2:91-8.
- Nailah S, Wan Ishak WMF, Essam A, Makky M. Comparison of Enzymes Production of Bacteria from Landfill Soil and Leachate: A Case Study-Jabor Landfill Kuantan, Pahang, Malaysia. Int J Innovation, Management and Technology 2014;5:1.
- Leonard DA, Bonomo RA, Powers RA. Class D ß-lactamases: a reappraisal after five decades. Accounts Chemical Research 2013;46:2407-15.
- Evans BA, Amyes SGB. OXA ß-Lactamases. Clinical Microbiology Rev 2014;27:241-63.
- Lagatolla C, Edalucci E, Dolzani L. Molecular evolution of metallo ß-lactamase producing Gram negative in a nosocomial setting of high-level endemicity. J Clin Microbiolo2006;44:2348-53.
- Chuang DM, Hough C, Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol 2005;45:269-s90.
- Kar N, Roy RN, Sen SK, Ghosh K. Isolation and characterization of extracellular enzyme-producing bacilli in the digestive tracts of rohu, Labeo rohita (Hamilton) and murrel, Channa punctatus. Asian Fish Sci 2008;21:421-34.
- Pandey A, Nigam P, Soccol CR. Advances in microbial amylases. Biotechnol Appl B Iochem 2000;31:135-52.
- Veerabagu M, Kirchler T, Elgass K. The interaction of the Arabidopsis response regulator ARR18 with bZIP63 mediates the regulation of proline dehydrogenase expression. Molecular Plant 2014;7:1560-77.
- Zhang YP, Hong J, Ye X. Cellulase assays Methods. Mol Biol 2009;581:213-1.
- Grabow WOK, Holtzhausen CS, de villiers JC. Research on Bacteriophages as Indicators of Water Quality. WRC Report No 321/1/93. Water Research Commission, Pretoria 1993;147.
- Usha K, Kumar E, Sai Gopal DVR. Occurrence of various beta-lactamase producing gram negative bacilli in the hospital effluent. Asian J Pharm Clin Res 2013;6:42.
- Nicoloff H, Andersson DI. Indirect resistance to several classes of antibiotics in cocultures with resistant bacteria expressing antibiotic-modifying or -degrading enzymes. J Antimicrob Chemother 2016;71:100-10.
- Albertson NH, Nystrom T, Kjelleberg S. Exoprotease activity of two marine bacteria during starvation. Appl Environ Microbiol 1990;56:218-23.
- Biswas J, Paul AK. Production of Extracellular Enzymes by Halophilic Bacteria Isolated from Solar Salterns. Int J Appl Biolo Pharmaceutical Technol 2013;4:30-6.
- Khalil S, Loyanchan TE, Tabatabai MA. Mycorrhizal dependency and nutrient uptake by improved and unimproved corn and soybean cultivars. Agron J 1994;86:949-58.
- Goñi-Urriza M, Capdepuy M, Arpin C. Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceaeand Aeromonas sp. Appl Environ Microbiol2000;66:125-32.
- Victor IB, David SW. Mechanisms of Antimicrobial Peptide Resistance in Gram-Negative Bacteria. Antibiotics 2015;4:18-41.
- Torres-Vila LM, Rodriguez Molina MC, Lacasa P. Pyrethroid resistance of Helicoverpa armigera in Spain: current status and agroecological perspective. Agriculture Ecosystems and Environment 2002;93:55-66.
- John H, Hatha AAM. Distribution, extracellular virulence factors and drug resistance of motile Aeromonads in fresh water ornamental fishes and associated carriage water. Int J Aquaculture 2013;3:92-100.